A HYBRID APPROACH FOR MALARIA CLASSIFICATION USING CNN-BASED FEATURE EXTRACTION AND TRADITIONAL MACHINE LEARNING CLASSIFIERS
Abstract
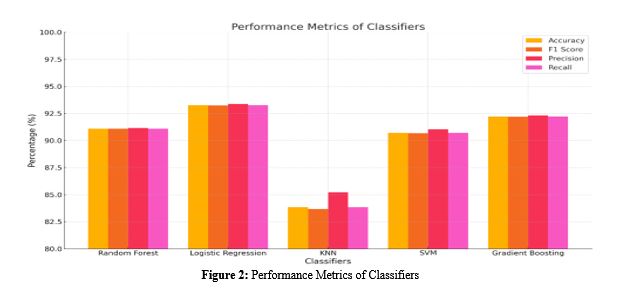
Malaria is a major global health threat, and timely and correct diagnosis is essential for effective treatment. Traditional diagnostic methods, such as the microscopic examination of blood smears, are time-consuming and require expert personnel. The study presents a mix of machine learning methods for automatic diagnosis of malaria by using the feature extraction capability of Convolutional Neural Networks (CNNs) along with the efficient classification performance of traditional machine learning classifiers. For our study, we utilize VGG16 CNN with a weight pre-trained on ImageNet to extract the features from non-infected and infected blood cell images from malaria. Five classical machine learning algorithms, such as Random Forest, Logistic Regression, K-Nearest Neighbours (KNN), Support Vector Machine (SVM), & Gradient Boosting, are used to classify the extracted features. Each classifier's performance is calculated based on accuracy, F1 score, precision, and recall metrics. The results of our experiments showed that the hybrid model has high accuracy in classification, where the Logistic Regression classifier could achieve above 93% accuracy. This hybrid method is a powerful diagnostic for malaria disease, accomplishing a more satisfactory compromise between the efficacy of the deep learning architectures such as CNNs, and the computational capabilities of more conventional classifiers. It holds promise for deployment into resource-limited settings where fast, automated threading diagnostic systems are much needed
Full text article
References
Abd Alnabi, D. L., Ahmed, S. S., & Abd Alnabi, N. L. (2025). knee osteoarthritis stage classification based on hybrid fusion deep learning framework. science journal of university of zakho, 13(2), 262-278. https://doi.org/10.25271/sjuoz.2025.13.2.1450
Ahamed, M. F., Nahiduzzaman, M., Mahmud, G., Shafi, F. B., Ayari, M. A., Khandakar, A., Abdullah-Al-Wadud, M., & Islam, S. M. R. (2025). Improving Malaria diagnosis through interpretable customized CNNs architectures. Scientific Reports, 15(1), 6484. https://doi.org/10.1038/s41598-025-90851-1
Al-Awadhi, M., Ahmad, S., & Iqbal, J. (2021). Current status and the epidemiology of malaria in the Middle East Region and beyond. Microorganisms, 9(2), 338. https://doi.org/10.3390/microorganisms9020338
Awosolu, O. B., Yahaya, Z. S., Farah Haziqah, M. T., & Olusi, T. A. (2022). Performance evaluation of nested polymerase chain reaction (Nested PCR), light microscopy, and plasmodium falciparum histidine-rich protein 2 rapid diagnostic test (PfHRP2 RDT) in the detection of falciparum malaria in a high-transmission setting in Southwestern Nigeria. Pathogens, 11(11), 1312. https://doi.org/10.3390/pathogens11111312
Balasubramaniam, S., Velmurugan, Y., Jaganathan, D., & Dhanasekaran, S. (2023). A modified LeNet CNN for breast cancer diagnosis in ultrasound images. Diagnostics, 13(17), 2746. https://doi.org/10.3390/diagnostics13172746
Das, R., Kumari, K., De, S., Manjhi, P. K., & Thepade, S. (2021). Hybrid descriptor definition for content based image classification using fusion of handcrafted features to convolutional neural network features. International Journal of Information Technology, 13(4), 1365–1374. https://doi.org/10.1007/s41870-021-00722-x
Gao, Z., Lu, Z., Wang, J., Ying, S., & Shi, J. (2022). A convolutional neural network and graph convolutional network-based framework for classification of breast histopathological images. IEEE Journal of Biomedical and Health Informatics, 26(7), 3163–3173. https://doi.org/10.1109/JBHI.2022.3153671
Grignaffini, F., Simeoni, P., Alisi, A., & Frezza, F. (2024). Computer-Aided Diagnosis Systems for Automatic Malaria Parasite Detection and Classification: A Systematic Review. Electronics, 13(16), 3174. https://doi.org/10.3390/electronics13163174
Litjens, G., Kooi, T., Bejnordi, B. E., Setio, A. A. A., Ciompi, F., Ghafoorian, M., Van Der Laak, J. A., Van Ginneken, B., & Sánchez, C. I. (2017). A survey on deep learning in medical image analysis. Medical Image Analysis, 42, 60–88. https://doi.org/10.1016/j.media.2017.07.005
Ramos-Briceño, D. A., Flammia-D’Aleo, A., Fernández-López, G., Carrión-Nessi, F. S., & Forero-Peña, D. A. (2025). Deep learning-based malaria parasite detection: convolutional neural networks model for accurate species identification of Plasmodium falciparum and Plasmodium vivax. Scientific Reports, 15(1), 3746. https://doi.org/10.1038/s41598-025-87979-5
Rosnelly, R., Riza, B. S., & Suparni, S. (2023). Comparative Analysis of Support Vector Machine and Convolutional Neural Network for Malaria Parasite Classification and Feature Extraction. https://doi.org/10.58346/JOWUA.2023.I3.015
Salehi, A. W., Khan, S., Gupta, G., Alabduallah, B. I., Almjally, A., Alsolai, H., Siddiqui, T., & Mellit, A. (2023). A study of CNN and transfer learning in medical imaging: Advantages, challenges, future scope. Sustainability, 15(7), 5930. https://doi.org/10.3390/su15075930
Shen, D., Wu, G., & Suk, H.-I. (2017). Deep learning in medical image analysis. Annual Review of Biomedical Engineering, 19(1), 221–248. https://doi.org/10.1146/annurev-bioeng-071516-044442
Authors
Copyright (c) 2025 Omar M. Ahmed, Walat A. Ahmed, Bafreen N. Mohammed, Asaad Kh. Ibrahim

This work is licensed under a Creative Commons Attribution 4.0 International License.
Authors who publish with this journal agree to the following terms:
- Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution License [CC BY-NC-SA 4.0] that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this journal.
- Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal's published version of the work, with an acknowledgment of its initial publication in this journal.
- Authors are permitted and encouraged to post their work online.