MOLECULAR DETECTION OF DRUG-RESISTANT GENES AMONG Clostridioides difficile FROM DIARRHEIC CHILDREN IN DUHOK CITY -IRAQ
Abstract
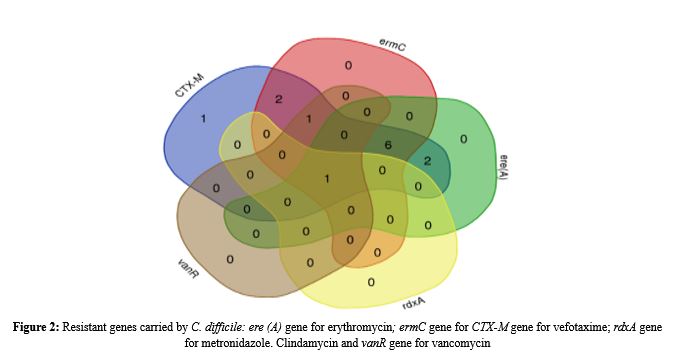
Clostridioides difficile, formerly known as Clostridium difficile is the most common cause of antibiotic-associated diarrhea and colitis and is characterized by resistance to multiple drugs. This study amid to characterize antibiotic resistance genes in C. difficile among pediatric diarrheal cases from Duhok Governorate, Kurdistan regional Iraq, providing critical insights for regional infection control and treatment guidelines. Thirteen C. difficile-positive stool samples (from a cohort of 200 children aged between 6 months- 6 years) were analysed by PCR for detecting resistance genes CTX-M (cefotaxime), ermC (clindamycin), ere(A) (erythromycin), rdxA (metronidazole), vanR (vancomycin). The result illustrated that cefotaxime, CTX-M gene detected in 100% DNA samples, with high rates of resistance of clindamycin (ermC gene, 76.9%) and erythromycin (69.2%, ere(A) gene) while resistance to metronidazole (rdxA) and vancomycin resistance (vanR) remained rare (7,69% and 15.38%, respectively). Venn diagram analysis highlighted frequent co-occurrence of resistance to these genes, and six samples (46.2%) harbored three genes, CTX-M, ermC and ere (A), and also double other two samples harbored two genes, CTX-M and ermC and CTX-M and ere (A). In addition, one sample harbored only the CTX-M gene. This study underscores the prevalence of the alarming high rate of antibiotic resistance found in C. difficile among pediatric diarrheal cases such as against cefotaxime, clindamycin, and erythromycin. The persistence of susceptibility to vancomycin and metronidazole supports their continued use as first-line therapies for both community and hospital infections region
Full text article
References
Al-Rawe, A. M., Yousif, Y. I., Al-Jomaily, O. K. G., Shaban, S. A., & Suleiman, A. A. (2023). Identification of Antimicrobial Resistance Genes and Drug Targets in Antibiotic-Resistant Clostridioides difficile Clinical Isolates. Molecular Genetics, Microbiology and Virology, 38(3), 197-206. https://DOI.org/10.3103/S0891416823030023
Baines, S. D., & Wilcox, M. H. (2015). Antimicrobial resistance and reduced susceptibility in Clostridium difficile: potential consequences for induction, treatment, and recurrence of C. difficile infection. Antibiotics, 4(3), 267-298. https://DOI.org/10.3390/antibiotics4030267
Baines, S. D., Noel, A. R., Huscroft, G. S., Todhunter, S. L., O'Connor, R., & Aktories, K. (2011). Evaluation of linezolid and rifaximin as treatment options for experimental Clostridium difficile infection. Journal of Antimicrobial Chemotherapy, 66(1), 133-139. DOI: 10.1093/jac/dkr155
Boekhoud, I. M., Nieuwenhuis, M., Knetsch, C. W., Kumar, N., Swart, R. L., Corver, J., & Kuijper, E. J. (2021). Antibiotic resistance of Clostridioides difficile isolates in the Netherlands, 2017-2020. Antimicrobial Resistance & Infection Control, 10(1), 1-10. DOI: 10.3390/pathogens11080949
Canas-Durán, R., Pérez-Segarra, O., Mendoza-Oliva, A., Gálvez, J., & Rodríguez-Diaz, J. (2023). Antibiotic Resistance in Clostridioides difficile: Current Situation and Future Prospects. Microorganisms, 11(2), 341. https://DOI.org/10.1016/bs.apcsb.2021.11.003
Carroll K, C, Bartlett J, G (2011). Biology of Clostridium difficile: implications for epidemiology and diagnosis. Ann Rev Microbiol 65: 501-521. doi: 10.1146/annurev-micro-090110- 102824. DOI: 10.1146/annurev-micro-090110-102824
Cassandra R., Duffy, Huang, Y., Andrikopoulou, M., Conrad, N., Asche, S., Jason D., Wright, N., Goffman, D., Mary, E., Alton, E., Alexander, M. F. (2020). Clindamycin, Gentamicin, and Risk for Clostridium difficile Infection and Acute Kidney Injury During Delivery Hospitalizations.Obstet Gynecol. 135(1): 59–67. DOI:10.1097/AOG.0000000000003568.
Debets-Ossenkopp, Y. J., Pot, R. G., Van Westerloo, D. J., Goodwin, A., Vandenbroucke-Grauls, C. M., Berg, D. E., ... & Kusters, J. G. (1999). Insertion of mini-IS 605 and deletion of adjacent sequences in the nitroreductase (rdxA) gene cause metronidazole resistance in Helicobacter pylori NCTC11637. Antimicrobial agents and chemotherapy, 43(11), 2657-2662. DOI: 10.1128/AAC.43.11.2657
Di, X., Bai, N., Zhang, X., Liu, B., Ni, W., Wang, J. & Wang, R. (2015). A meta-analysis of metronidazole and vancomycin for the treatment of Clostridium difficile infection, stratified by disease severity. Brazilian Journal of Infectious Diseases, 19, 339-349.DOI: 10.1016/j.bjid.2015.03.006
Dupuy, B., Govind, R., Antunes, A., & Matamouros, S. (2006). Clostridium difficile toxin synthesis is negatively regulated by TcdC. Journal of Medical Microbiology, 57(6), 685-689.DOI: 10.1099/jmm.0.47775-0
Ghaffar NM, Mohialdeen NH. Isolation and molecular characterization of Campylobacter jejuni from local broiler chicken (lbc) and frozen imported chickens (ifc) in duhok province, kurdistan region-iraq. Science Journal of University of Zakho. 2023 Aug 10; 11(3):386-95. DOI: 10.1099/jmm.0.47775-0
Hami, Iman A., and Khalid S. Ibrahim. "Incidence of methicillin-resistant Staphylococcus aureus (MRSA) recovered from patients with urinary tract infections in Zakho City/ Kurdistan-Iraq." Science Journal of University of Zakho 11.1 (2023): 91-97. DOI: https://DOI.org/10.25271/sjuoz.2023.11.1.1041
Hasan SM, Ibrahim KS. Molecular characterization of extended-spectrum β-lactamase (ESBL) and virulence gene factors in uropathogenic Escherichia coli (UPEC) in children in Duhok City, Kurdistan Region, Iraq. Antibiotics. 2022 Sep 14; 11(9):1246. DOI: 10.3390/antibiotics11091246
Ibrahim D. R. (2023). Prevalence of Plasmid Mediated QNRA, QNRB and QNRS Among Clinical Escherichia Coli Isolated from Urinary Tract Infections in Duhok, Kurdistan Region of Iraq. Science Journal of the University of Zakho. 11(4):523-31. https://DOI.org/10.25271/sjuoz.2023.11.4.1196
Issa F. A. (2024). Antibiotic Resistance Patterns of Common Uropathogens Isolated from Females at Zakho City, Kurdistan Region, Iraq. Science Journal of the University of Zakho.12(4):490-506. https://DOI.org/10.25271/sjuoz.2024.12.4.1395
Khashei R, Malekzadegan Y, Ebrahim-Saraie HS, Razavi Z. (2018). Phenotypic and genotypic characterization of macrolide, lincosamide and streptogramin B resistance among clinical isolates of staphylococci in southwest of Iran. BMC Res. Notes 11: 711. DOI: 10.1186/s13104-018-3817-4
Kikuchi, E., Miyamoto, Y., Narushima, S., & Itoh, K. (2002). Design of Species‐specific primers to identify 13 species of Clostridium harbored in human intestinal tracts. Microbiology and immunology, 46(5), 353-358. DOI: 10.1111/j.1348-0421.2002.tb02706.x
Lessa, F. C., Mu, Y., Bamberg, W. M., Beldavs, Z. G., Dumyati, G. K., Dunn, J. R.and Fridkin, S. K. (2015). Burden of Clostridium difficile infection in the United States. New England Journal of Medicine, 372(9), 825-834. DOI: 10.1056/NEJMoa1408913.
Mehdi L. Y and AL-Mossawei M. T. (2015). PCR for detection of Clostridium difficile toxin A (tcdA) and toxin B (tcdB) genes in Iraq. Journal of Health, Medicine and Nursing 87: 57-68.
Miele, A., Bandera, M., & Goldstein, B. P. (1995). Use of primers selective for vancomycin resistance genes to determine van genotype in enterococci and to study gene organization in VanA isolates. Antimicrobial agents and chemotherapy, 39(8), 1772-1778. DOI: 10.1128/aac.39.8.1772
Mohialdeen N. H and Ghaffar N. M. (2023). Isolation and molecular characterization of Campylobacter jejuni from local broiler chicken (lbc) and frozen imported chickens (ifc) in duhok province, kurdistan region-iraq. Science Journal of University of Zakho. 11(3):386-95. https://DOI.org/10.25271/sjuoz.2023.11.3.1136
Ofosu, A. (2016). Clostridium difficile infection: a review of current and emerging therapies. Annals of gastroenterology: quarterly publication of the Hellenic Society of Gastroenterology, 29(2), 147. DOI: 10.20524/aog.2016.0006
Owens J, R. C., Donskey, C. J., Gaynes, R. P., Loo, V. G., & Muto, C. A. (2008). Antimicrobial-associated risk factors for Clostridium difficile infection. Clinical Infectious Diseases, 46(Supplement_1), S19-S31. https://DOI.org/10.1086/521859
Saeed A.Y. and Ibrahim, K. S. (2013). Detection of Enterohemorrhagic Escherichia Coli O157 in Sheep and Goats Using Fluorogenic and Chromogenic Culture Media. Science Journal of University of Zakho. 1(1):115-9.
Sidjabat, H. E., Paterson, D. L., Adams-Haduch, J. M., Ewan, L., Pasculle, A. W., Muto, C. A., ... & Doi, Y. (2009). Molecular epidemiology of CTX-M-producing Escherichia coli isolates at a tertiary medical centre in western Pennsylvania. Antimicrobial agents and chemotherapy, 53(11), 4733-4739. DOI: 10.1128/AAC.00533-09
Sun, X., & Hirota, S. A. (2015). The roles of host and pathogen factors and the innate immune response in the pathogenesis of Clostridium difficile infection. Molecular immunology, 63(2), 193-202. DOI: 10.1016/j.molimm.2014.09.005
Sun, X., & Hirota, S. A. (2015). The roles of host and pathogen factors and the innate immune response in the pathogenesis of Clostridium difficile infection. Molecular immunology, 63(2), 193-202. DOI: 10.1016/j.molimm.2014.09.005
Taher F. S. and Othman H. E. (2024). Molecular identification and genotyping of methicillin-resistant staphylococcus aureus (mrsa) in different clinical samples. Science Journal of University of Zakho. 12(2):159-68.https://DOI.org/10.25271/sjuoz.2024.12.2.1276
Tamma, P. D., Antel, A. S., Avdic, E., Carroll, K. C., Mikolajczak, A., Natarajan, K., ... & Simner, P. J. (2022). Clostridium difficile infection: epidemiology, diagnosis, and antimicrobial susceptibility testing. Journal of Clinical Microbiology, 60(6), e00518-22. DOI: 10.1038/nrgastro.2016.25
Tao, S., Chen, H., Li, N., Wang, T., & Liang, W. (2022). The spread of antibiotic resistance genes in vivo model. Canadian Journal of Infectious Diseases and Medical Microbiology, 2022(1), 3348695. DOI: 10.1155/2022/3348695
Van, T. T. H., Chin, J., Chapman, T., Tran, L. T., and Coloe, P. J. (2008). Safety of raw meat and shellfish in Vietnam: an analysis of Escherichia coli isolations for antibiotic resistance and virulence genes. International journal of food microbiology, 124(3), 217-223. DOI: 10.1016/j.ijfoodmicro.2008.03.029
Yahav, D., Koay, T. H., Karnik, N. D., & Adler, A. (2023). Antimicrobial resistance in Clostridioides difficile infection: A narrative review. Antimicrobial Resistance & Infection Control, 12(1), 1-12. DOI: 10.1186/s13756-020-00815-5
Authors
Copyright (c) 2025 Bakhtyar Nader Ali

This work is licensed under a Creative Commons Attribution 4.0 International License.
Authors who publish with this journal agree to the following terms:
- Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution License [CC BY-NC-SA 4.0] that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this journal.
- Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal's published version of the work, with an acknowledgment of its initial publication in this journal.
- Authors are permitted and encouraged to post their work online.