AN EXPLAINABLE DEEP LEARNING FRAMEWORK FOR BREAST CANCER CLASSIFICATION USING EFFICIENTNETV2B0 AND GRAD-CAM
Abstract
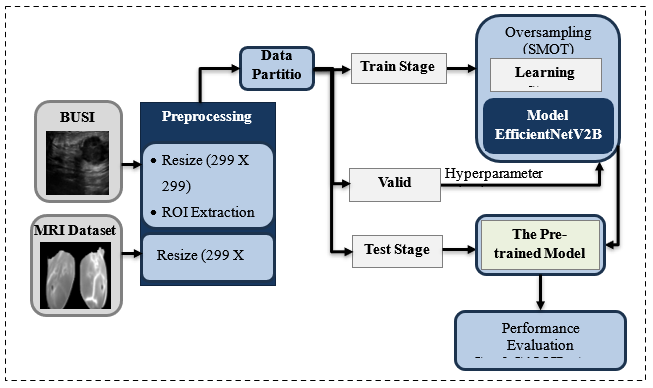
Breast cancer remains one of the most serious health challenges worldwide, withearly and accurate diagnosis can significantly improve patient outcomes. Traditional diagnostic methods often rely heavily on expert interpretation, which may lead to inconsistencies or delays in decision-making. To address this issue, this research provides a deep-learning framework that uses the EfficientNetV2B0 model in combination with Grad-CAM (Gradient-weighted Class Activation Mapping) to provide illustrated explanations to detect breast cancer using ultrasound and MRI datasets. Our method addresses serious challenges such as class imbalance and irrelevant image characteristics by employing SMOTE (Synthetic Minority Over-sampling Technique) oversampling and Region of Interest (ROI) extraction for BUSI (Breast Ultrasound Images) datasets. The Grad-CAM approach improves reliability and transparency by providing visual proof that support’s each decision, allowing healthcare professionals to better understand the AI's prediction. Trained and assessed on two different medical imaging datasets, the framework obtained extraordinarily high accuracy (98.97% on BUSI and 99.55% on MRI), along with low prediction error and high reliability. The model is both accurate and understandable, making it ideal for clinical usage. It is also faster and more dependable than current approaches, making it highly beneficial.
Full text article
References
Abioye, O. A., Evwiekpaefe, A. E., & Awujoola, A. J. (2024). PERFORMANCE EVALUATION OF EFFICIENTNETV2 MODELS ON THE CLASSIFICATION OF HISTOPATHOLOGICAL BENIGN BREAST CANCER IMAGES. Science Journal of University of Zakho, 12(2), 208–214. https://doi.org/10.25271/sjuoz.2024.12.2.1261
Adadi, A., & Berrada, M. (2018). Peeking inside the black-box: a survey on explainable artificial intelligence (XAI). IEEE Access, 6, 52138–52160.
Akram, M., Iqbal, M., Daniyal, M., & Khan, A. U. (2017). Awareness and current knowledge of breast cancer. Biological Research, 50, 1–23.
Albahri, A. S., Duhaim, A. M., Fadhel, M. A., Alnoor, A., Baqer, N. S., Alzubaidi, L., Albahri, O. S., Alamoodi, A. H., Bai, J., & Salhi, A. (2023). A systematic review of trustworthy and explainable artificial intelligence in healthcare: Assessment of quality, bias risk, and data fusion. Information Fusion, 96, 156–191.
Al-Dhabyani, W., Gomaa, M., Khaled, H., & Fahmy, A. (2020). Dataset of breast ultrasound images. Data in Brief, 28. https://doi.org/10.1016/j.dib.2019.104863
Arnold, M., Morgan, E., Rumgay, H., Mafra, A., Singh, D., Laversanne, M., Vignat, J., Gralow, J. R., Cardoso, F., Siesling, S., & Soerjomataram, I. (2022). Current and future burden of breast cancer: Global statistics for 2020 and 2040. Breast, 66, 15–23. https://doi.org/10.1016/j.breast.2022.08.010
Bhushan, A., Gonsalves, A., & Menon, J. U. (2021). Current state of breast cancer diagnosis, treatment, and theranostics. Pharmaceutics, 13(5), 723.
Botchkarev, A. (2018). Performance metrics (error measures) in machine learning regression, forecasting and prognostics: Properties and typology. ArXiv Preprint ArXiv:1809.03006.
Brian J Burkett, C. W. H. (2016). A Review of Supplemental Screening Ultrasound for Breast Cancer: Certain Populations of Women with Dense Breast Tissue May Benefit. Academic Radiology.
Chai, T., & Draxler, R. R. (2014). Root mean square error (RMSE) or mean absolute error (MAE). Geoscientific Model Development Discussions, 7(1), 1525–1534.
Chawla, N. V, Bowyer, K. W., Hall, L. O., & Kegelmeyer, W. P. (2002). SMOTE: synthetic minority over-sampling technique. Journal of Artificial Intelligence Research, 16, 321–357.
Cruz-Ramos, C., García-Avila, O., Almaraz-Damian, J. A., Ponomaryov, V., Reyes-Reyes, R., & Sadovnychiy, S. (2023). Benign and Malignant Breast Tumor Classification in Ultrasound and Mammography Images via Fusion of Deep Learning and Handcraft Features. Entropy, 25(7). https://doi.org/10.3390/e25070991
Dessain, J. (2022). Machine learning models predicting returns: Why most popular performance metrics are misleading and proposal for an efficient metric. Expert Systems with Applications, 199, 116970.
Dianpei Ma, C. W. J. L. X. H. Y. Z. Z. G. C. L. C. L. Y. H. (2025). Analysis of the diagnostic efficacy of ultrasound, MRI, and combined examination in benign and malignant breast tumors. Frontiers in Oncology.
Hassija, V., Chamola, V., Mahapatra, A., Singal, A., Goel, D., Huang, K., Scardapane, S., Spinelli, I., Mahmud, M., & Hussain, A. (2024). Interpreting black-box models: a review on explainable artificial intelligence. Cognitive Computation, 16(1), 45–74.
Jabeen, K., Khan, M. A., Hamza, A., Albarakati, H. M., Alsenan, S., Tariq, U., & Ofori, I. (2024). An EfficientNet integrated ResNet deep network and explainable AI for breast lesion classification from ultrasound images. CAAI Transactions on Intelligence Technology. https://doi.org/10.1049/cit2.12385
Jeni, L. A., Cohn, J. F., & De La Torre, F. (2013). Facing imbalanced data--recommendations for the use of performance metrics. 2013 Humaine Association Conference on Affective Computing and Intelligent Interaction, 245–251.
Krithiga, R., & Geetha, P. (2021). Breast cancer detection, segmentation and classification on histopathology images analysis: a systematic review. Archives of Computational Methods in Engineering, 28(4), 2607–2619.
Luong, H. H., Nguyen, H. T., & Thai-Nghe, N. (2024). AEGANB3: An Efficient Framework with Self-attention Mechanism and Deep Convolutional Generative Adversarial Network for Breast Cancer Classification. In IJACSA) International Journal of Advanced Computer Science and Applications (Vol. 15, Issue 5). www.ijacsa.thesai.org
Mersha, M., Lam, K., Wood, J., AlShami, A., & Kalita, J. (2024). Explainable artificial intelligence: A survey of needs, techniques, applications, and future direction. Neurocomputing, 128111.
Nasir, M. U., Ghazal, T. M., Khan, M. A., Zubair, M., Rahman, A. U., Ahmed, R., Hamadi, H. Al, & Yeun, C. Y. (2022). Breast Cancer Prediction Empowered with Fine-Tuning. Computational Intelligence and Neuroscience, 2022. https://doi.org/10.1155/2022/5918686
Nieto-Castanon, A., Ghosh, S. S., Tourville, J. A., & Guenther, F. H. (2003). Region of interest based analysis of functional imaging data. Neuroimage, 19(4), 1303–1316.
Pathan, R. K., Alam, F. I., Yasmin, S., Hamd, Z. Y., Aljuaid, H., Khandaker, M. U., & Lau, S. L. (2022). Breast Cancer Classification by Using Multi-Headed Convolutional Neural Network Modeling. Healthcare (Switzerland), 10(12). https://doi.org/10.3390/healthcare10122367
Podda, A. S., Balia, R., Barra, S., Carta, S., Fenu, G., & Piano, L. (2022). Fully-automated deep learning pipeline for segmentation and classification of breast ultrasound images. Journal of Computational Science, 63. https://doi.org/10.1016/j.jocs.2022.101816
Rasheed, K., Qayyum, A., Ghaly, M., Al-Fuqaha, A., Razi, A., & Qadir, J. (2022). Explainable, trustworthy, and ethical machine learning for healthcare: A survey. Computers in Biology and Medicine, 149, 106043.
Sahu, A., Das, P. K., & Meher, S. (2024). An efficient deep learning scheme to detect breast cancer using mammogram and ultrasound breast images. Biomedical Signal Processing and Control, 87. https://doi.org/10.1016/j.bspc.2023.105377
Selvaraju, R. R., Cogswell, M., Das, A., Vedantam, R., Parikh, D., & Batra, D. (2020). Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. International Journal of Computer Vision, 128(2), 336–359. https://doi.org/10.1007/s11263-019-01228-7
Tan, M., & Le, Q. V. (2021). EfficientNetV2: Smaller Models and Faster Training. https://github.com/google/
Uzair Khan. (2021). Breast cancer patients MRI’s. Kaggle.
Vigil, N., Barry, M., Amini, A., Akhloufi, M., Maldague, X. P. V., Ma, L., Ren, L., & Yousefi, B. (2022). Dual-Intended Deep Learning Model for Breast Cancer Diagnosis in Ultrasound Imaging. Cancers, 14(11). https://doi.org/10.3390/cancers14112663
Waks, A. G., & Winer, E. P. (2019). Breast cancer treatment: a review. Jama, 321(3), 288–300.
Wang, J. Z. (2001). Integrated region-based image retrieval (Vol. 11). Springer Science & Business Media.
Zhang, J., Zhang, Z., Liu, H., & Xu, S. (2023). SaTransformer: Semantic-aware transformer for breast cancer classification and segmentation. IET Image Processing, 17(13), 3789–3800. https://doi.org/10.1049/ipr2.12897
Zuluaga-Gomez, J., Al Masry, Z., Benaggoune, K., Meraghni, S., & Zerhouni, N. (2021). A CNN-based methodology for breast cancer diagnosis using thermal images. Computer Methods in Biomechanics and Biomedical Engineering: Imaging and Visualization, 9(2), 131–145. https://doi.org/10.1080/21681163.2020.1824685
Authors
Copyright (c) 2026 Jane M. Haj Ali and Diman Hassan

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.
Authors who publish with this journal agree to the following terms:
- Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution License [CC BY-NC-SA 4.0] that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this journal.
- Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal's published version of the work, with an acknowledgment of its initial publication in this journal.
- Authors are permitted and encouraged to post their work online.