WEIGHTED GENE CO-EXPRESSION NETWORK ANALYSIS IDENTIFIES BIOLOGICAL PATHWAYS AND BIOMARKER GENES ASSOCIATED WITH CHICKENS' ADAPTATION TO BOTH LOW AND HIGH ALTITUDES
Abstract
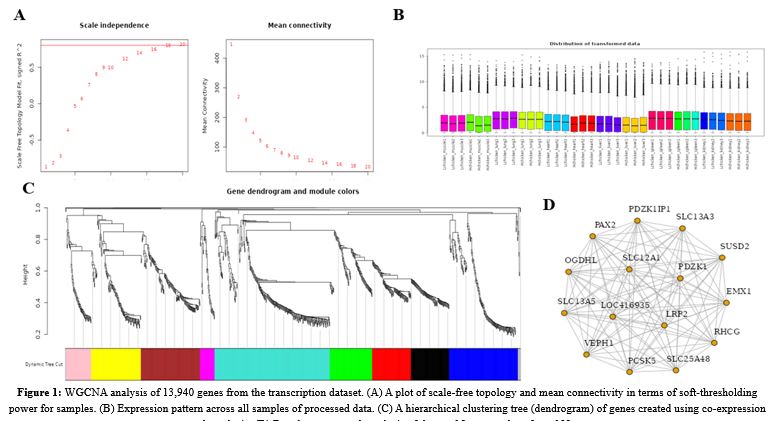
The main aim of the study was to identify modules, hub genes, and possible pathways linked with hypoxia adaptation in six types of tissues and organs (heart, kidney, liver, lung, muscle, and spleen) at altitudes ranging from 2,300 to 3,500 metersOn a transcription dataset from hypoxia-sensitive tissues, we performed weighted gene co-expression network analysis on 13,940 selected genes, and 10 transcriptional modules in total were detected (Turquoise 196 genes, Purple 27 genes, Blue 196, Brown 182, Yellow 108, Green 79 genes, Red 69 genes, Black 50 genes, Pink 44 genes, and Magenta 37 genes). Furthermore, we discovered that the majority of variable genes were screened by sub-setting 1000 genes; samples belonging to the same tissue clearly clustered together, and the expression in the liver and lung was more associated than in the heart and spleen. Functional enrichment analysis of all genes in 12 selected modules revealed that 9 KEGG pathways were considerably enriched, 13 Gene ontology terms were significantly enriched in the biological process and cellular component pathways, and 15 gene ontology terms were significantly enriched in the molecular function pathway. Through weighted gene co-expression network analysis, the results of this study expand our knowledge of the molecular pathway of catalytic and metabolic activity as a biomarker pathway
Full text article
References
Anderson, C. M., & Stahl, A. (2013). SLC27 fatty acid transport proteins. Molecular Aspects of Medicine, 34(2), 516–528. DOI: https://doi.org/10.1016/j.mam.2012.07.010
Bao, Q., Zhang, X., Bao, P., Liang, C., Guo, X., Chu, M., & Yan, P. (2021). Using weighted gene co-expression network analysis (WGCNA) to identify the hub genes related to hypoxic adaptation in yak (Bos grunniens). Genes & Genomics, 43(10), 1231–1246. DOI: https://doi.org/10.1007/s13258-021-01137-5
Cheng, J., Sun, Y., Zhang, X., Zhang, F., Zhang, S., Yu, S., Qiu, X., Tan, L., Song, C., Gao, S., Wu, Y., & Ding, C. (2014). Toll-like receptor 3 inhibits Newcastle disease virus replication through activation of pro-inflammatory cytokines and the type-1 interferon pathway. Archives of Virology, 159(11), 2937–2948. DOI: https://doi.org/10.1007/s00705-014-2148-6
Cheviron, Z. A., & Brumfield, R. T. (2012). Genomic insights into adaptation to high-altitude environments. Heredity, 108(4), 354–361. DOI: https://doi.org/10.1038/hdy.2011.85
Fu, Z., Akula, S., Olsson, A.-K., & Hellman, L. (2022). Chicken cathepsin G-like - A highly specific serine protease with a peculiar tryptase specificity expressed by chicken thrombocytes. Developmental & Comparative Immunology, 129, 104337. DOI: 10.1016/j.dci.2021.104337
Ge, S. X., Son, E. W., & Yao, R. (2018). iDEP: an integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinformatics, 19(1), 534. DOI: https://doi.org/10.1186/s12859-018-2486-6
Heidari, M., Sarson, A. J., Huebner, M., Sharif, S., Kireev, D., & Zhou, H. (2010). Marek’s disease virus-induced immunosuppression: array analysis of chicken immune response gene expression profiling. Viral Immunology, 23(3), 309–319. DOI: 10.1089/vim.2009.0079
Kang, Y., Feng, M., Zhao, X., Dai, X., Xiang, B., Gao, P., Li, Y., Li, Y., & Ren, T. (2016). Newcastle disease virus infection in chicken embryonic fibroblasts but not duck embryonic fibroblasts is associated with elevated host innate immune response. Virology Journal, 13(1), 41. DOI: https://doi.org/10.1186/s12985-016-0499-1
Langfelder, P., & Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics, 9(1), 559. DOI: https://doi.org/10.1186/1471-2105-9-559
Li, S., Zhang, X., Dong, X., Guo, R., Nan, J., Yuan, J., Schlebusch, C. M., & Sheng, Z. (2023). Genetic structure and characteristics of Tibetan chickens. Poultry Science, 102(8), 102767. DOI: https://doi.org/10.1016/j.psj.2023.102767
Matulova, M., Varmuzova, K., Sisak, F., Havlickova, H., Babak, V., Stejskal, K., Zdrahal, Z., & Rychlik, I. (2013). Chicken innate immune response to oral infection with Salmonella enterica serovar Enteritidis. Veterinary Research, 44(1), 37. DOI: 10.1186/1297-9716-44-37
O’Brien, K. A., Simonson, T. S., & Murray, A. J. (2020). Metabolic adaptation to high altitude. Current Opinion in Endocrine and Metabolic Research, 11, 33–41. DOI: https://doi.org/10.1016/j.coemr.2019.12.002
Papandreou, I., Cairns, R. A., Fontana, L., Lim, A. L., & Denko, N. C. (2006). HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metabolism, 3(3), 187–197. DOI: 10.1016/j.cmet.2006.01.012
Ranaware, P. B., Mishra, A., Vijayakumar, P., Gandhale, P. N., Kumar, H., Kulkarni, D. D., & Raut, A. A. (2016). Genome Wide Host Gene Expression Analysis in Chicken Lungs Infected with Avian Influenza Viruses. PLOS ONE, 11(4), e0153671. DOI: https://doi.org/10.1371/journal.pone.0153671
Rue, C. A., Susta, L., Cornax, I., Brown, C. C., Kapczynski, D. R., Suarez, D. L., King, D. J., Miller, P. J., & Afonso, C. L. (2011). Virulent Newcastle disease virus elicits a strong innate immune response in chickens. Journal of General Virology, 92(4), 931–939. DOI: https://doi.org/10.1099/vir.0.025486-0
S., D. M., A., G. R., A., B. D., R., K. T., M., D. J. C., Huaijun, Z., & J., L. S. (2017). Novel Mechanisms Revealed in the Trachea Transcriptome of Resistant and Susceptible Chicken Lines following Infection with Newcastle Disease Virus. Clinical and Vaccine Immunology, 24(5), e00027-17. DOI: https://doi.org/10.1128/CVI.00027-17
Schilling, M. A., Katani, R., Memari, S., Cavanaugh, M., Buza, J., Basu, J. R., Mpenda, F. N., Deist, M. S., Lamont, S. J., & Kapur, V. (2018). Transcriptional innate immune response of the developing chicken embryo to Newcastle disease virus infection. Frontiers in Genetics, 9(FEB), 1–9. DOI: 10.3389/fgene.2018.00061
Sha, Y., Gao, C., Liu, M., & Zhao, S. (2020). Evaluation of the genetic diversity of six Chinese indigenous chickens. Asian-Australasian Journal of Animal Sciences, 33(10), 1566–1572. DOI: 10.5713/ajas.19.0606
Shen, Y., Zhang, J., Yang, J., Liu, C., Bian, S., Zhang, C., Yu, J., Gao, X., & Huang, L. (2020). Association of EPAS1 and PPARA Gene Polymorphisms with High-Altitude Headache in Chinese Han Population. BioMed Research International, 2020. DOI: 10.1155/2020/1593068
Tang, Q., Gu, Y., Zhou, X., Jin, L., Guan, J., Liu, R., Li, J., Long, K., Tian, S., Che, T., Hu, S., Liang, Y., Yang, X., Tao, X., Zhong, Z., Wang, G., Chen, X., Li, D., Ma, J., … Li, M. (2017). Comparative transcriptomics of 5 high-altitude vertebrates and their low-altitude relatives. GigaScience, 6(12), 1–9. DOI: 10.1093/gigascience/gix105
Team, R. C. (2014). R: A language and environment for statistical computing. MSOR Connections, 1. DOI: https://api.semanticscholar.org/CorpusID:215755663
Wang, Y., Zhu, Q., Yang, L., & Liu, Y. P. (2012). Ontogenic expression pattern and genetic polymorphisms of the fatty acid transport protein 4 (FATP4) gene in Chinese chicken populations. International Journal of Molecular Sciences, 13(6), 6820–6835. DOI: 10.3390/ijms13066820
Authors
Copyright (c) 2025 Pshtiwan S.A. Bebane, Paiman Yousif, Sarbast I. Mustafa, Sami mamand

This work is licensed under a Creative Commons Attribution 4.0 International License.
Authors who publish with this journal agree to the following terms:
- Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution License [CC BY-NC-SA 4.0] that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this journal.
- Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal's published version of the work, with an acknowledgment of its initial publication in this journal.
- Authors are permitted and encouraged to post their work online.